Interaction with Pandas Package¶
Warning
You’ll need to install Pandas first.
Pandas is a powerful data analysis package that provides the user a large set of functionalities, such as easy slicing, filtering, calculating and summarizing statistics or plotting.
Python-dwca-reader exposes a pd_read() method to easily load the content of a data file (core or extension)
from the archive into a Pandas DataFrame.
from dwca.read import DwCAReader
with DwCAReader('gbif-results.zip') as dwca:
print("Core data file is: {}".format(dwca.descriptor.core.file_location)) # => 'occurrence.txt'
core_df = dwca.pd_read('occurrence.txt', parse_dates=True)
# All Pandas functionalities are now available on the core_df DataFrame
Note
DwCAReader.pd_read() is a simple wrapper around
pandas.read_csv() and accept the same
optional arguments. Only a few of them (delimiter, skiprows, encoding, …) will be ignored because DwCAReader
sets them appropriately for the data file.
Note
Alternatively, you can do core_df = dwca.pd_read(dwca.core_file_location, …) which is handy if you don’t know the name of the core data file.
As a small example, some applications on the core_df:
Warning
You’ll need to install Seaborn for this example.
import pandas as pd
import seaborn as sns
# Number of records for each institutioncode
core_df["institutionCode"].value_counts()
# Select the coordinate information of the first twenty records
core_df.loc[:20, ["decimalLatitude", "decimalLongitude"]]
# Count the number of records with date information after 1950
sum(core_df["year"] > 1950)
# Convert eventDate to DateTime python object
core_df['eventDate'] = pd.to_datetime(core_df['eventDate'])
# Select only those records with coordinates, not (0, 0) coordinates and eventDate provided
core_df[(core_df["decimalLatitude"] != 0.0) &
(core_df["decimalLatitude"].notnull()) &
(core_df["decimalLongitude"] != 0.0) &
(core_df["decimalLongitude"].notnull()) &
(core_df["eventDate"].notnull())]
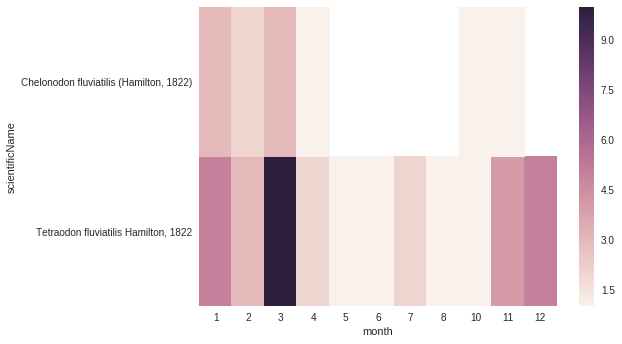
# Count the number of records for each species for each month
count_occ = core_df.pivot_table(index="scientificName",
columns="month",
values="id",
aggfunc='count')
# Visualisation of the counts on a heatmap (Seaborn)
sns.heatmap(count_occ)
For more information about Pandas and Seaborn, see their respective documentation.
When the DwCA contains multiple files, joining the extensions with the core file could be of interest for further analysis.
import pandas as pd
from dwca.read import DwCAReader
with DwCAReader('dwca-2extensions.zip') as dwca:
# Check the core file of the Archive (Occurrence, Taxon, ...)
print("Core type is: {}".format(dwca.descriptor.core.type))
# Check the available extensions
print("Available extensions: {}".format([ext.split("/")[-1] for ext in dwca.descriptor.extensions_type]))
taxon_df = dwca.pd_read('taxon.txt')
descr_df = dwca.pd_read('description.txt')
vern_df = dwca.pd_read('vernacularname.txt')
# Join the information of the description and vernacularname extension to the core taxon information
# (cfr. database JOIN)
taxon_df = pd.merge(taxon_df, descr_df, left_on='id', right_on='coreid', how="left")
taxon_df = pd.merge(taxon_df, vern_df, left_on='id', right_on='coreid', how="left")
The result is the core file joined with the extension files. More information about the Pandas merge is provided in the documentation.
Remark that reading in the data to Pandas will load the entire file into memory. For large archives, this won’t be
feasible. Pandas support the usage of chunks, reading in a processing the data in chunks. As an example, consider the
selection of those occurrences for which the eventDate was a Sunday:
import pandas as pd
from dwca.read import DwCAReader
chunksize = 10 # Chosen chunksize to process the data (pick a larger value for real world cases)
with DwCAReader('gbif-results.zip') as dwca:
sunday_occ = []
for chunk in dwca.pd_read('occurrence.txt', chunksize=chunksize):
chunk['eventDate'] = pd.to_datetime(chunk['eventDate'])
# Subselect only the records recorded on a sunday
sunday_occ.append(chunk[chunk['eventDate'].dt.weekday == 6]) # Monday = 0, Sunday = 6
sunday_occ = pd.concat(sunday_occ)
More advanced processing is supported by Pandas. However, when only interested in counting the number of occurrences for
a specific condition, Pandas is not always required. As an example, counting the number of occurrences for each species
in the data set is easily supported by the Counter datatype of Python:
from collections import Counter
from dwca.read import DwCAReader
from dwca.darwincore.utils import qualname as qn
with DwCAReader('/Users/nicolasnoe/Desktop/gbif-results.zip') as dwca:
count_species = Counter()
for row in dwca:
count_species.update([row.data[qn('scientificName')]])
print(count_species)
Hence, the added value of Pandas depends on the type of analysis. Some more extensive applications of Pandas to work with Darwin Core data is provided in this data cleaning tutorial and data analysis tutorial.